High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Craven, G.B., Affron, D.P., Allen, C.E., Matthies, S., Greener, J.G., Morgan, R.M.L., Tate, E.W., Armstrong, A., Mann, D.J.(2018) Angew Chem Int Ed Engl 57: 5257-5261
- PubMed: 29480525
- DOI: https://doi.org/10.1002/anie.201711825
- Primary Citation of Related Structures:
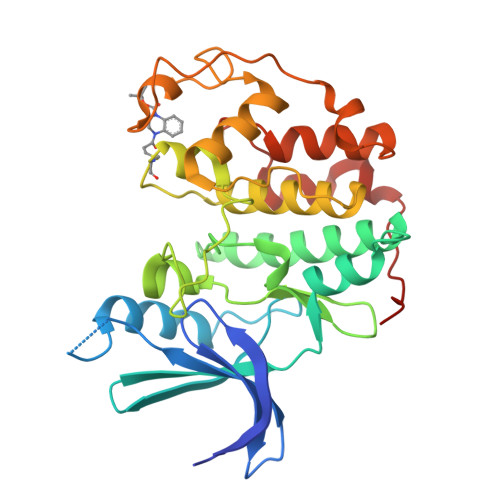
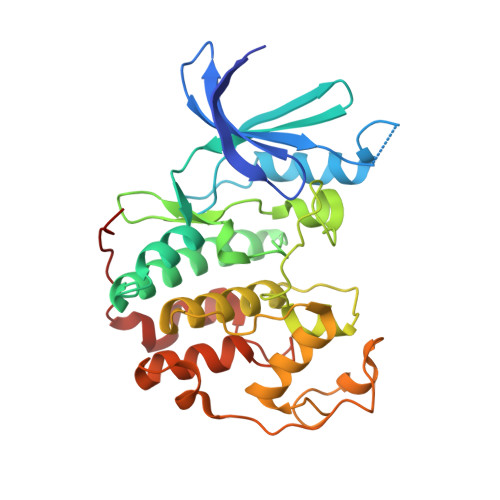
5OO0, 5OSJ, 5OSM - PubMed Abstract:
Cysteine-reactive small molecules are used as chemical probes of biological systems and as medicines. Identifying high-quality covalent ligands requires comprehensive kinetic analysis to distinguish selective binders from pan-reactive compounds. Quantitative irreversible tethering (qIT), a general method for screening cysteine-reactive small molecules based upon the maximization of kinetic selectivity, is described. This method was applied prospectively to discover covalent fragments that target the clinically important cell cycle regulator Cdk2. Crystal structures of the inhibitor complexes validate the approach and guide further optimization. The power of this technique is highlighted by the identification of a Cdk2-selective allosteric (type IV) kinase inhibitor whose novel mode-of-action could be exploited therapeutically.
Organizational Affiliation:
Department of Chemistry, Imperial College London, South Kensington Campus, London, SW7 2AZ, UK.